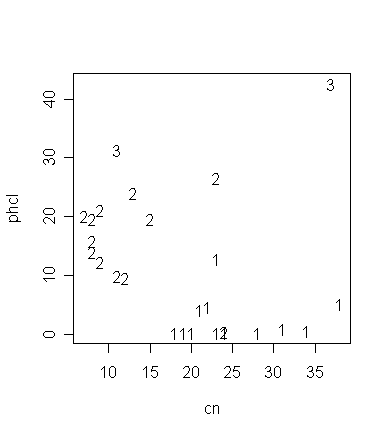
わかりやすくするために R を使って解析した例を紹介します。変数 cn、phcl の値とサンプルのグループ帰属 が group で以下のように示されているとします。
cn phcl group
[1,] 34 0.5 1
[2,] 28 0.2 1
[3,] 24 0.2 1
[4,] 23 0.3 1
[5,] 19 0.3 1
[6,] 20 0.2 1
[7,] 24 0.4 2
[8,] 23 26.6 2
[9,] 13 24.1 2
[10,] 15 19.7 2
[11,] 38 5.1 1
[12,] 21 4.2 1
[13,] 12 9.6 2
[14,] 18 0.3 1
[15,] 23 12.9 1
[16,] 31 1.0 1
[17,] 37 42.6 3
[18,] 8 19.6 2
[19,] 7 20.2 2
[20,] 9 21.1 2
[21,] 8 14.0 2
[22,] 8 15.8 2
[23,] 9 12.3 2
[24,] 11 31.4 3
[25,] 22 4.6 1
[26,] 11 9.9 2
|
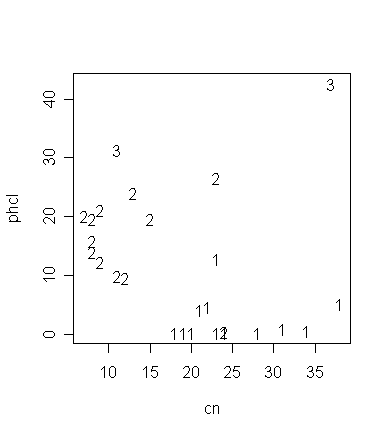
> library(MASS)
> z <- lda(group ~ cn + phcl)
> z
Call:
lda(group ~ cn + phcl)
Prior probabilities of groups:
1 2 3
0.46153846 0.46153846 0.07692308
Group means:
cn phcl
1 25.08333 2.483333
2 12.25000 16.108333
3 24.00000 37.000000
Coefficients of linear discriminants:
LD1 LD2
cn 0.05613327 -0.13082789
phcl -0.16064435 -0.05322542
Proportion of trace:
LD1 LD2
0.8536 0.1464
|
> x <- cbind(cn, phcl)
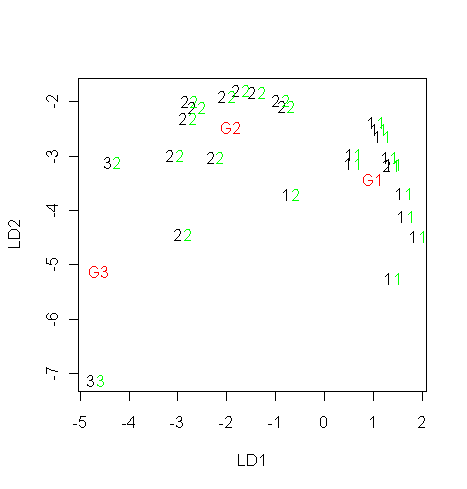
> z$scaling # 正準変量固有ベクトル
LD1 LD2
cn 0.05613327 -0.13082789
phcl -0.16064435 -0.05322542
> u <- x %*% z$scaling # 正準変量平面への変換
> plot(u, type="n")
> text(u, as.character(group))
> um <- z$means %*% z$scaling # 正準変量平面上でのグループ平均
> text(um, c("G1","G2","G3"), col="red")
> pg <- predict(z)
> up <- u
> up[,1] <- up[,1]+0.2
> text(up, as.character(pg$class), col="green") # グループ帰属を緑で表示
|